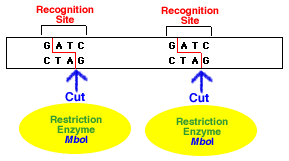
Restriction enzymes have the ability to cut DNA molecules at very precise sequences of 4-8 base pairs called recognition sites. They naturally occur in many bacteria as a defence mechanism against bacteriophages (viruses that infect bacteria). The restriction enzymes in the bacteria were able to cut the viral DNA, usually at least at several locations, thus rendering the viral DNA inactive so it couldn't infect the bacterium.
Purified forms of these restriction enzymes are now used as tools to cut DNA. Scientists use over 400 restriction enzymes that recognize roughly 100 recognition sites as a means to isolate, sequence and manipulate individual genes taken from any cell (including humans).
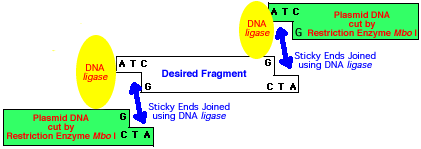
The sites where the fragments of DNA are cut may result in overhanging "sticky ends" (See diagrams below.) Pieces with similar sticky ends can be joined together using the enzyme DNA ligase. Hybrid DNA from different sources (or species) may be produced this way and such DNS is known as recombinant DNA.
1. A restriction enzyme cuts the double-stranded DNA molecule at its specific recognition site.



Transgenesis